Journal of Vaccines and Immunology
Avian influenza
Ebtsam M El-Kady1*, Hanaa A El-Shafei2 and SoadA Saleh1
2Microbial chemistry Department, National Research Centre, Cairo, Egypt
Cite this as
El-Kady EM, El-Shafei HA, Saleh S (2019) Avian influenza. J Vaccines Immunol 5(1): 022-027. DOI: 10.17352/jvi.000026Influenza A H5N1 subtype known as avian flu is an emerging and extremely contagious virulent disease that poses a risk to the international community’s security. In latest years, by establishing itself as a important emerging infectious disease, it has become a major public health issue.
It is one of more than 25 influenza A viruses that mainly live in all bird species [1]. It has been noted that migratory birds can play a part in avian influenza virus transmission, black-tailed gull and chickens infection with avian influenza (H13N2) and (H13N8) viruses in eastern China. They discovered these H13 viruses transmitted to national poultry from migratory birds.
Avian flu is infecting mammals and animals as well. The primary source of human infections is direct contact with infected poultry or contaminated environments [2]. Influenza A virus may be categorized as avian influenza, swine influenza, or other animal influenza viruses, depending on the hostsource . Examples include subtypes A(H9N2) and A(H5N1)of avian influenza “bird flu” or subtypes A(H3N2) and A(H1N1) of swine influenza virus. All these Type A animal influenza viruses are different from human influenza viruses and do not spread readily among animals. Highly pathogenic avian influenza (HPAI) is named poultry viruses that cause severe disease and result in elevated mortality levels. Low pathogenic avian influenza (LPAI) is called viruses in poultry that cause mild disease [3].
Following unprotected interaction with infected birds or avian influenza virus contaminated surfaces, human infections with some avian viruses happened most often. However, some infections were recognized where there was no known direct contact. The disease of people varied from mild to severe.
Currently, recognized avian influenza viruses do not readily communicate from individual to individual, the continuing circulation of these viruses in poultry is disturbing as these viruses cause serious disease in animals and have the ability to mutate to become more contagious among animals
The main risk factor for human disease, such as live bird markets, tends to be direct or indirect exposure to infected live or dead poultry or polluted environment, In terms of human risk factors for avian influenza virus infections. Often involved are slaughtering, beating, handling infected carcasses of poultry and preparing poultry for consumption, particularly in household environment.
There are four types of influenza viruses as reported by WHO (2018), types A-D: Influenza A infects people and livestock. An influenza pandemic can be caused by the creation of a fresh influenza A virus capable of infecting people and maintaining human transmission.
Influenza B viruses cause seasonal epidemics and circulate among animals. It is also possible to infect seals. Scientists reported that H3N8 a type of bird flu is responsible for the death of the more than 160 harbor seals on the East Coast [4].
Influenza C viruses can infect individuals and pigs, but infections are usually mild and seldom reported. Influenza D viruses primarily affect bovine animals and are not known to affect human beings or cause disease.
Form A strains of influenza: Influenza A has the greatest genetic variation and the largest range of host [5].
Influenza viruses are listed further by their genes of Neuraminidase (NA) and Hemagglutinin (HA) into subtypes, there are 18 HA and 11 NA kinds as of now.
Considerable progress has been produced towards knowing the fundamental foundation for the association of the two major influenza A viruses with their dominant ligand/substrate surface glycoproteins: sialic acid-ended carbohydrate chains Heamagglutinin mediates target cell attachment virus specificity, which in its receptor binding site acquires distinctive modifications to alter.
Hemagglutinin (HA)
Influenza virus HA protein binds the virus particle to the sensitive cell receptor.
It is the main antigen where antibodies are directed to neutralize.
The ongoing development of fresh strains and subsequent influenza epidemics is mainly accountable for this.
HA, agglutinates red blood cells, the foundation of a diagnostic test called the inhibition heamagglutination test.
Neuraminidase
To release progeny virus from infected cells, it cleaves neuramic acid. In the respiratory tract, the protective layer of mucus is also degraded. This increases the ability of the virus to penetrate the respiratory epithelium. Influenza viruses, particularly influenza A viruses, demonstrate constant changes in their protein antigenicity of heamagglutinin and neuraminidase, and to a lower degree in type B, where type C appears to be antigenically stable.
Influenza A Viruses (IAV) may exchange genetic material in co-infected cells during a reassortment phase [6].
Two forms of antigenic modifications occur
Major changes are taking place based on the reassortment of RNA genome sections. During reassortment, whole pieces of RNA are exchanged, each coding for a fresh protein such as hemagglutinin. Most animal species have their influenza A virus, such as marine birds, ducks, swine and horses, which is the cause of RNA sections encoding epidemic antigenic variations.
Antigenic shifts
There are major changes based on the reassortment of segments of the RNA genome. In reassortment, entire segments of RNA are exchanged, each one of which codes for a new protein e.g. hemagglutinin. Many species of animals e.g. aquatic birds, chickens, swine and horses, have their influenza A virus which is the source of the RNA segments that encode the antigenic shift variants of epidemics among humans.
Antigenic drifts
These are minor changes based on the genome RNA mutations.
High rates of mutation
Influenza viruses have a comparatively large mutation rate typical of RNA viruses. Its genome segmentation promotes genetic recombination in hosts simultaneously infected with two distinct influenza viruses by segment reassortment [7,8].
A previously uncontagious strain, one of several possible routes to a pandemic, can then pass between individuals. The capacity to demonstrate species-selectivity of different influenza strains are mainly due to hemagglutinin gene variation.
Genetic mutations that cause single amino acid replacements in the hemagglutinin gene can significantly change the ability of viral hemagglutinin proteins to bind receptors on the surface of the host cells. These improvements in avian H5N1 viruses may alter the inefficiency of virus strains as more prevalent kinds of human influenza virus [9]. This does not imply that a replacement of one amino acid will cause a pandemic, but it does mean that this replacement can allow a strain of avian flu to become pathogenic in animals.
Influenza virus replication cycle
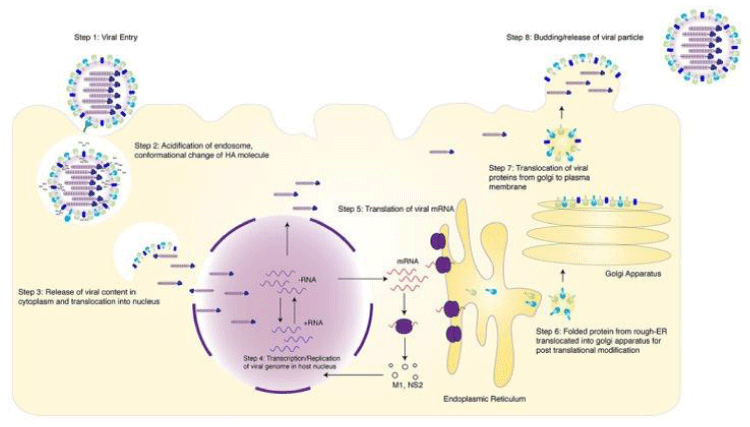
The replication process of influenza relies on single stranded segmented genomes that encode 10 viral proteins (M1, M2, NA, HA,NS1, NP, NEP, PA,PB1 and PB2) [10]. These proteins are formed by 8 segmented NP-coated genomic strands [11], have a double-helical hairpin structure, and bear one heterotrimer of polymerase composed of PB1, PB2 and PA (particles of the ribonucleoprotein). Figure 1 shows the different steps of replication cycle.
Step 1: The HA protein on the surface of the virion acknowledges and binds to sialic acid on the host cell surface after cell entry via Lakadamyali [12], receptor mediated endocytosis, the endosome passes into the cell and changes the pH more acidically [13]. As a consequence, the acidification method creates an irreversible conformational change to the influenza virus in the HA molecule and exposes the peptide of hydrophobic fusion [14]. Inserting the peptide of fusion into the endosomal membrane that causes the bacterial and endosomal membrane to fuse [15].
Step 2: In the tetramer where the M2 channel serves as a pore [16-18]. The M2 protein acts as an ion channel that modulates the intravirion’s pH, injecting free hydrogen atoms into the viral nucleus, dissociating the vRNPs from the M1 matrix proteins [16,19].
Step 3: The earlier mechanism enables the inner viral core material to be released into the cytoplasm and inserted into the nucleus [12,16].
Step 3,4: Upon dissociation from M1, VRNPs are moved to the host nucleus, where viral replication and transcription happens [12,16].
Step 4: Influenza viruses are among the few species with RNA that can replicate in a host core due to a cap sequence required to transcribe RNA polymerase [20].
Step 5,6: Influenza viral RNA sections missing a 5 cap for transcription of RNA-dependent RNA polymerase, so the PA ,PB1 and PB2, elements conduct cap-snatching of host DNA to finish this process (20-22) Cap-containing viral mRNA is discharged through the host ribosome apparatus into the cytoplasm to bedeciphered. Surface proteins such as NA and HA are changed over into the rough endoplasmic reticulum and translocated for post-translation adjustments into the Golgi apparatus [23].
Step 7: The NS2 viral atomic send out protein is basic to the atomic trade of vRNPs, which has resulted in decreased viral growth [24], without NS2. A protein synthesis NS2 and M1 assist guide the fresh viral proteins to the host cell film after genomic replication, transcription where they gather and bud crisply synthesized virions [25].
Step 8: As the host cell’s nascent virions grow, NA cleaves the host cell membrane’s sialic acid residues [26], permitting the viruses to elude the host layer.
Antiviral drugs that block NA activity result in influenza viruses accumulating at the membrane and cannot furthur disseminate to cause infection of neighbouring cells. Antiviral drugs that square NA action result in membrane-accumulating influenza viruses and are unable to spread to cause nearby cell infection [27].
Infection mechanism
In human instances of avian influenza infection, the main mode of transmission is from bird to human by direct or near contact with infected birds or contaminated surfaces. Christine and Jiang (2008) reported that H5N1 avian influenza is an extremely deadly irresistible malady that lead to possibly catastrophic widespread in case the H5N1 virus changes effectively among humans, taints separated aspiratory epithelial cells and makes diffuse alveolar hurt and dying within the lungs of tainted patients. The infection may too taint other organs, counting the trachea ,guts and brain ,and may enter the placental obstruction, passing through pregnant woman placenta to infect the fetus.
Signs and symptoms in humans
Avian, swine, and other human zoonotic flu maladies can cause ailment extending from gentle upper respiratory disease (fever and hack) to quick movement to genuine pneumonia, stun and indeed death.
Pandemic potential
Influenza pandemics are epidemics caused by a novel virus affecting a big percentage of the globe. Pandemics are eccentric but repeating occasions that can have a worldwide wellbeing, financial, and social impact. When a unused flu infection emerges with the capacity to cause persistent human-to-human transmission, a flu widespread happens and the human population has small or low resistance [28].
Epidemiology of human infections
Since the beginning of the 20th century, there have been four influenza widespread each triggered by the appearance of a new infection. The source of the influenza virus responsible for the pandemic of 1918, which in a single year killed more individuals than the bubonic plague, continues unsure, but it appears to have been an adapted avian influenza strain. The new viruses contained in the pandemics of 1957 and 1968.
Human HPAI A(H5N1) virus infections were recorded during a poultry epidemic in China ,Hong Kong and Saudi Arabia , in 1997. Since 2003 this avian infection appeared in Asia and spread to Europe and Africa and has gotten to be endemic in some countrie’s poultry populaces. A fresh pandemic began in North America in 2009 when a novel H1N1 human-swine-avian reassortment virus emerged. By the end of 2017 , H5N1 influenza virus resulted in 454 deaths (WHO).
The emergence of a novel H1N1 human-swine-avian reassortment virus in 2009 in North America started a new pandemic. At the end of 2017, 860 laboratory confirmed cases of H5N1 influenza virus infection from 16 different countries, resulting in 454 deaths had been reported to the World Health Organization (WHO).
Flare-ups have driven in millions of illnesses of poultry, hundred cases of people and many fatilities. In influenced countries, flare-ups of poultry have extremely influenced employments, the economy and the worldwide trade.
Human A(H7N9) virus infections were first recorded in China in 2013. The virus has since spread across the nation in the poultry population, resulting in more than 1,500 recorded human instances and many fatalities.
Other avian influenza viruses have led to sporadic diseases of humans, including A(H7N7) and A(H9N2). A few countries too recorded scattered human diseases with swine flu infection, especially subtypes A(H3) and A(H1).
Diagnosis
It is essential for all laboratory staff to be prepared and well informed about the laboratory techniques used to diagnose avian influenza due to the elevated concern concerning Type A influenza, particularly avian influenza. Through their spit, nasal discharges, feces and blood, contaminated winged creatures convey H5N1. Other animals may gotten to be tainted with the infection by coordinate contact with these body liquids or by contact with sullied surfaces [29].
In arrange to analyze human disease with zoonotic flu, research facility tests are required. Through its worldwide flu reconnaissance and reaction framework the WHO routinly overhauls specialized direction conventions for zoonotic flu recognizable proof in people utilizing molecular for case. Strategies of RT-PCR and others. Rapid Influenza Diagnostic Tests (RIDTs) have decreased affectability than PCR and their unwavering quality is basically subordinate beneath which they are utilized. Subtype information cannot generally be provided by commercially accessible RDTs. RIDTs are sometimes used in clinical environments, but they are restricted in their use to detect zoonotic viruses.
Treatment
Evidence indicates that certain antiviral drugs, particularly inhibitors of neuraminidase (oseltamivir, zanamivir), may decrease the length of viral replication and enhance survival prospects. Neuraminidase inhibitors ought to be endorsed as rapidly as conceivable in suspected and confirmed instances (ideally within 48 hours of the start of symptoms) to maximize restorative focal point. Be that as it may, given the vital motality directly related with subtype infection contaminations of A(H7N9) and A(H5) and confirmation of amplified viral replication in these sicknesses, the medicate ought to too be administered.
Treatment is recommended for at least 5 days, but may be expanded till there is sufficient clinical improvement. Except for other purposes (e.g., asthma and other specific conditions) corticosteroids should not be used routinely; as associated with excessive viral clearance, immunosuppressant leading to bacterial or fungal super infection. The latest A(H7N9) and A(H5) viruses are antiviral resistant to adamantane (e.g., amantadine and rimantadine) and are not suggested for monotherapy.In critically ill patients, co-infection with bacterial pathogens may occur.
Prevention
In addition to antiviral therapy, the management of public health includes personal protective measures such as: Regular hand washing and proper hand drying. Good respiratory hygiene-covering the mouth and nose while coughing or sneezing, using and disposing of tissues properly. Early self-isolation of those who feel unwell, feverish, and other influenza symptoms. Avoid close contact with patients. Avoid touching their face, nose, or mouth. If possible, travelers to nations and individuals residing in nations with known avian influenza outbreaks should avoid contact with poultry farms, livestock on live poultry markets, and any surfaces that appear to be contaminated with poultry or other animal feces.
Vaccines
Immunization has been one of the most conclusive advances prompting the drastic descending pattern in the frequency of numerous viral illnesses. The aim of immunization is to induce a “primed” state in the vaccinated subject (killed virus or live attenuated virus) so that a rapid secondary immune response is generated after being exposed to a pathogen triggering the quickened eradication organism and clinical disease prevention. Achievement depends on the production of T and B cells for memory and on the presence of an antibody in the serum.
Influenza Vaccine
Flu infection displays a significant and relentless threat to general wellbeing around the world, and current vaccinations give immunity to viral secludes like the immunization strain. High-affinity antibodies to a controlled epitope can give different flu subtypes insusceptibility and assurance against possible pandemic infections. Co-crystal structures have been resolved at 2.2 and 2.7 angstrom resolutions to extensively suppress human immune response CR6261 Fab in complexes with main surface antigen (hemagglutinin, HA) from 1918 H1N1 flu pandemic. and an ongoing deadly instance of H5N1 avian flu. CR6261 perceives a closely regulated helical locale in the membrane-proximal stem of HA1 and HA2 in constant to other structurally identified flu antibodies. Recognized here, the CR6261 epitope will facilitate the development and execution of enhanced vaccines that can elicit CR6261-like antibodies as well as immunizer-based flu treatments [30].
Binding influenza hemagglutinin (HA) and cell receptor sialic acids cause flu diseases. To discharge the flu genome into the cytoplasm, the binding is followed by camouflage, endocytosis and un-covering. It is conceivable that the replication of flu viruses could be reversed by unique inhibitors that strange any of these cases.
Rehashed flu infection contaminations are normal due to the fast antigenic variety of glycoproteins in the viral envelope.Host infection is avoided by antibodies to the viral neuraminidase and hemagglutinin proteins. Nonetheless, new vaccines, including antigens derived from flu strains now circulating in the population, are distributed each year as a result of the rapid antigenic range. At present, identification of flu strains allows for the selection of sufficient antigens for each season. The antibodies consist of partly purified proteins from existing A and B strains of inactivated influenza.
Making oligopeptides as antigenic determinants
The most defined way to construct a vaccine would be to manufacture the antigenic determinants. Including relevant B and T cell epitopes and arrange them in a properly large molecule. Doing so, it would be possible to snake an assembly where the interesting epitopes are suitably organized to elicit an optimal immune response. To synthesized T cell epitopes seems quite feasible since they generally are composed of specific amino acid sequences. It is more difficult to synthesize B cell epitopes, which often are dependent on so called conformational epitopes, which often consist of adjacent amino acid residues located discontinuously on the peptide chain. However, in some cases an oligopeptides seems to be adequately immunogenic as a B cell epitopes provided it is supplemented with relevant T cell epitopes. In most cases the B cell epitopes (oligopeptides) are cross-linked to a carrier molecule. The efficacy of cross-linked is crucial, since a sufficient number of epitopes are needed per carrier molecule for optimal presentation to elicit a high antibody response to the epitope [31].
In fact, there is an additional structural dimension to make peptides optimally immunogenic as subunits in comparison to protein molecules, the reason being that, first, an efficient coupling method has to be worked out to conjugate the peptide to the carrier, second, the conjugate has to be formulated into multimeric complexes like a micelle, virosomes, or an lscom.Using biotin as a small molecule. studied the density of biotin molecules required to make biotin immunogenic.
Recombinant (re-) baculoviruses (BV) methodology has filled in as a suitable alternative for the high all through, high fidelity expression of numerous novel discharged human genes. Until this point in time, more than 75 human genes have been expressed, and the re-protein decontaminated. This expression framework consolidates numerous great attributes including relative speed, moderate expense but most essentially, the making of naturally active proteins [32].
Immunology of the conventional and rDNA vaccines
How does immune response initiation occur?
There are proteins labeled “antigen” in the disease-causing species that activate the immune response. The resulting immune response is multifold and involves protein production called “antibodies.” Such proteins bind to the pathogens and ultimately destroy them. In addition, in an immune response, “memory cells” are formed. These are cells that remain in the circulation system, some of the time for the host’s life span, ready to mount a rapid defensive immune response to follow-up infections with the same disease-causing agent that triggered their development. Furthermore, “memory cells” are formed in an immune response. These are cells that remain in the circulation system, some time for the life span of the host, eager to mount a rapid defensive immune response to follow-up infections with the same disease-causing agent that triggered their growth.
Avian influenza A viruses (IAVs) normally contaminate distinctive avian species, and auqatic birds. Once in a while, avian IAVs, for example, H5N1 and H7N9 infections can be transmitted to human, causing severe illness. Antigenically novel avian flu infections that contaminate and cause sickness in people represent a potential pandemic danger in the event that they can spread productively from individual to individual. The immune response of the host is critical in deciding infection pathogenesis and is the reason for the improvement of control techniques. In this review, we analyze the intrinsic and versatile immune responsiveness to avian influenza viruses and their role in disease and recovery. Besides, we examine the advancement in creating immunizations against avian IAVs and abridge hindrances in structuring all-inclusive and pandemic flu vaccines [33]. The only preventive measure to control flu is vaccine. Inactivated vaccines are currently the main source for prophylaxis of influenza. Usually, they are prepared from an infection produced in embryonic eggs of chicken, isolated from the allantoic liquids of hatched eggs, and inactivated using formaldehyde or β-propiolactone to formulate whole-virus antibody.
Alternatively, for broken or subunit vaccine formulation, the diluted virus is treated with ether or detergent. These inactivated immunizations are then intramuscularly or subcutaneously incubated into humans. Nevertheless, the high pathogenicity of the now-circling H5N1 infections raises vaccine structure challenges. HPA1 H5N1 virus can not be used as a seed virus for inactivated immunization because it not only jeopardizes the lives of antibody producers, but also makes it more difficult to obtain high-caliber allantoic fluid with a worthy virustiters from embryonated eggs. Subsequently, successful H5N1 vaccinations are desperately needed. Inactivated vaccines based on reverse genetics have been developed in compliance with WHO proposals and are now undergoing clinical evaluation in a few countries.
- Li Z, Tang T, Wu W, Gu L, Du J (2016) Efficacy of nasal continuous positive airway pressure on patients with OSA with erectile dysfunction and low sex hormone levels. Respir Med 119: 130-134. Link: http://bit.ly/2PUNaEJ
- Edler AA (2006) Avian flu (H5N1): its epidemiology, prevention, and implications for anesthesiology. J Clin Anesth 18: 1-4. Link: http://bit.ly/32rL1mG
- ElMasry I, Elshiekh H, Abdlenabi A, Saad A, Arafa A, et al. (2017) Avian influenza H5N1 surveillance and its dynamics in poultry in live bird markets, Egypt. Transbound Emerg Dis 64: 805-814. Link: http://bit.ly/2JSeeRp
- Kinneen L (2012) Bird Flu Not Responsible For Alaska Ice Seal, Walrus Illness. Link: http://bit.ly/33rm1NJ
- Gamblin SJ, Skehel JJ (2010) Influenza Hemagglutinin and Neuraminidase Membrane Glycoproteins. J Biol Chem 285: 28403-28409. Link: http://bit.ly/2WWgEUj
- White MC, Tao H, Steel J, Lowen AC (2019) H5N8 and H7N9 packaging signals constrain HA reassortment with a seasonal H3N2 influenza A virus. Proc Natl Acad Sci 116: 4611-4618. Link: http://bit.ly/33mZpOw
- World Health Organization. Global influenza program surveillance network (2005) Evolution of H5N1 avian influenza viruses in Asia. Emerg Infect Dis 11: 1515-1521. Link: http://bit.ly/2NncFgk
- Kou Z, Lei FM, Yu J, Fan ZJ, Yin ZH, et al. (2005) New genotype of avian influenza H5N1 viruses isolated from tree sparrows in China. J Virol 79: 15460-15466. Link: http://bit.ly/32rMk54
- Gambaryan A, Tuzikov A, Pazynina G, Bovin N, Balish A, et al. (2006) Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology 344: 432-438. Link: http://bit.ly/2NQY1Nx
- Flint SJ, Racaneillo VR, Rall GF, Skalka AM (2015) Principles of virology, Fourth Edition Bundle 569. Link: http://bit.ly/32rxThk
- McGeoch D, Fellner P, Newton C (1976) Influenza virus genome consists of eight distinct RNA species. Proc Natl Acad Sci U S A 73: 3045-3049. Link: http://bit.ly/34xk9mG
- Wu WW, Sun YH, Panté N (2007) Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol J 4: 49. Link: http://bit.ly/2Q0Voeu
- Lakadamyali M, Rust MJ, Zhuang X (2004) Endocytosis of influenza viruses. Microbes Infect 6: 929-936. Link: http://bit.ly/2pERBsT
- Hu YB, Dammer EB, Ren RJ, Wang G (2015) The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener 4: 18. Link: http://bit.ly/2ChuNlB
- White J, Kielian M, Helenius A (1983) Membrane fusion proteins of enveloped animal viruses. Q Rev Biophys 16: 151-195. Link: http://bit.ly/2qyfx0M
- Martin K, Helenius A (1991) Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol 65: 232-244. Link: http://bit.ly/2NooPp9
- Holsinger LJ, Nichani D, Pinto LH, Lamb RA (1994) Influenza A virus M2 ion channel protein: a structure-function analysis. J Virol 68: 1551-1563. Link: http://bit.ly/2PVNzqw
- Cross TA, Dong H, Sharma M, Busath DD, Zhou HX (2012) M2 Protein from influenza A: from multiple structures to biophysical and functional insights. Curr Opin Virol 2: 128-133. Link: http://bit.ly/34v6wEG
- Helenius A (1992) Unpacking the incoming influenza virus. Cell 69: 577-578. Link: http://bit.ly/34J4GAh
- Ulmanen I, Broni BA, Krug RM (1981) Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m(7)GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci USA 78: 7355-7359. Link: http://bit.ly/2CiKo4g
- Blaas D, Patzelt E, Kuechler E (1982) Identification of the cap binding protein of influenza virus. Nucleic Acids Res 10: 4803-4812. Link: http://bit.ly/36FEPLh
- Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, et al. (2008) The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature 289: 500-506. Link: http://bit.ly/34xlxpo
- Yadav V, Panganiban AT, HonerZu, Bentrup K, Voss TG (2016) Influenza infection modulates vesicular trafficking and induces Golgi complex disruption. Virus disease 27: 357-368. Link: http://bit.ly/2JWZLn3
- O’Neill RE, Talon J, Palese P (1998) The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J 17: 288-296. Link: http://bit.ly/2JWUuMt
- Whittaker G, Bui M, Helenius A (1996) The role of nuclear import and export in influenza virus infection. Trends Cell Biol 6: 67-71. Link: http://bit.ly/2JWRiQY
- Gottschalk A (1957) Neuraminidase: the specific enzyme of influenza virus and Vibrio cholerae. Biochim Biophys Acta 23: 645-646. Link: http://bit.ly/2NpMUfq
- McKimm-Breschkin JL (2013) Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respir Viruses 7: 25-36. Link: http://bit.ly/2oYNOpU
- Neumann G, Noda T, Kawaoka Y (2009) Emergence and pandemic potential of swine-origin H1N1 influenza virus Nature 459: 931-939. Link: http://bit.ly/2WR420F
- Alexander DJ (2008) Avian influenza - diagnosis. Zoonoses Public Health 55: 16-23. Link: http://bit.ly/36CAAjr
- Ekiert DC1, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, et al. (2009) Antibody recognition of a highly conserved influenza virus epitope. Science 324: 246-251. Link: http://bit.ly/2PVZtke
- Lovgren K., Lindmark J, Pipkorn R, Morein B (1987) Antigenic presentation of small molecules and peptide conjugated to a preformed iscom as carrier. J Immunol Methods 98: 137-143. Link: http://bit.ly/2PTLknJ
- Coleman TA, Parmelee D, Thotakura NR, Nguyen N, Bürgin M, et al. (1997) Production and purification of novel secreted human proteins. Gene 190: 63-171. Link: http://bit.ly/2qur8hw
- Koutsakos M, Kedzierska K, Subbarao K (2019) Immune Responses to Avian Influenza Viruses. J Immunol 202: 382-391. Link: http://bit.ly/32swqqZ
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
