Annals of Antivirals and Antiretrovirals
Case study – apply a deep-learning algorithm to exomes detection with online resources
Maria da Conceição Proença1* and António Pedro Alves de Matos2
2Centre for Interdisciplinar Research Egas Moniz (CiiEM), Monte da Caparica, Portugal
Cite this as
Conceição Proença MD, Alves de Matos AP (2021) Case study – apply a deep-learning algorithm to exomes detection with online resources. Ann Antivir Antiretrovir 5(1): 033-035. DOI: 10.17352/aaa.000014Copyright
© 2021 Conceição Proença MD, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Exosomes are membrane vesicles that constitute a potential mode of intercellular communication. Although the scope of its role is still being discussed, the release of exosomes by tumor cells suggests their participation in pathological situations and its study will surely drive to new therapeutic and diagnostic strategies. It is therefore important to be able to localize and identify these kinds of particles in transmission electron microscopy images. This preliminary work shows how a recent deep-learning algorithm available online can be applied to exosomes images to localize all the instances of the objects of interest, in any scale and several backgrounds.
Introduction
Exosomes are membrane vesicles that are released by cells upon fusion of multivesicular bodies with the plasma membrane, and measure up to 30 to 100, 120, or 150 nm according to the authors [1-3] that are perfectly observed with Transmission Electron Microscopy (TEM) images, although the use of nanoparticle tracking analysis is being suggested lately. The interest in exosomes recently increase [4] exponentially, with the discovery they carry mRNA and microRNA with a role in pathological processes and development in health disorders of several types, such as viral, hereditary and neoplastic diseases [5] and have a potential value as biomarkers or therapeutic tools.
Material and methodology
The image dataset came from the Centre for Biomedical Image Analysis at Masaryk University, Czech Republic. The exosomes were isolated by ultracentrifugation from the ascites of ovarian cancer patients, negatively contrasted with ammonium molybdate, and imaged with a Morgagni 268D microscope (FEI) equipped with Megaview III (Soft Imaging System), at 70 kV.
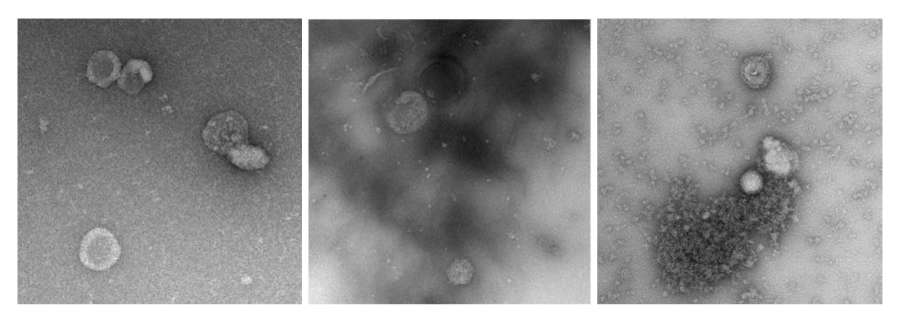
The dimensions of the individual images are 500x500 pixels, with scales ranging from 1.0 to 2.5 nanometers per pixel. There is also some diversity of the backgrounds, as several images are completely smooth, while others contain structures of different sizes and appearances (Figure 1) that are not exosomes.
The YOLOv5 algorithm can be downloaded and installed and will run from a command window in Windows-based systems. To work on our data, we need to annotate two subsets of images, one for train and another for validation. Usually, around 30% of the images available will be allocated to train and validation, split into 20-10%. Both subsets must be annotated: annotation is the equivalent of ground truth in the terminology of supervised classification, a set of images where the objects of interest are identified by the class and localized.
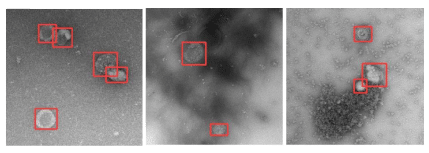
Nowadays, all procedures can be done with tools available online, such as [9]. After choosing a set of representative images, we uploaded it and proceed to draw boxes around the objects of interest in each image; at the end of the process, a file in appropriate format to be read by YOLO v5 is exported, and it contains all the labels we’ve been drawing, so all we have to do is distribute the images and the files of labels between two directories with fixed names that the training procedure will recognize.
There are a few parameters for train options and a set of hyperparameters that allow for data augmentation – variations in intensity, scale, rotation, shear, reflections, etc. that can be applied to the data available to increase the spectra of the objects in the training stage. The train can take many hours, depending mainly on the model used – a model x with a Convolutional Neural Network (CNN) of 607 layers will take more time for each iteration than a model s, with just 283 layers, the size of the images, and the regularization method.
When the train ends the number of iterations demanded (here called epochs), a series of graphic outputs assess the performance of the train/validation procedure based on precision (% of true positives correctly classified) and recall (% of true positives correctly detected among all positives). The file resulting from the train can now be used for inference. Any image of the same kind can now be submitted to the detector and evaluated in milliseconds, the result being the identification and localization of the objects of interest (Figure 2) always with the same criterium.
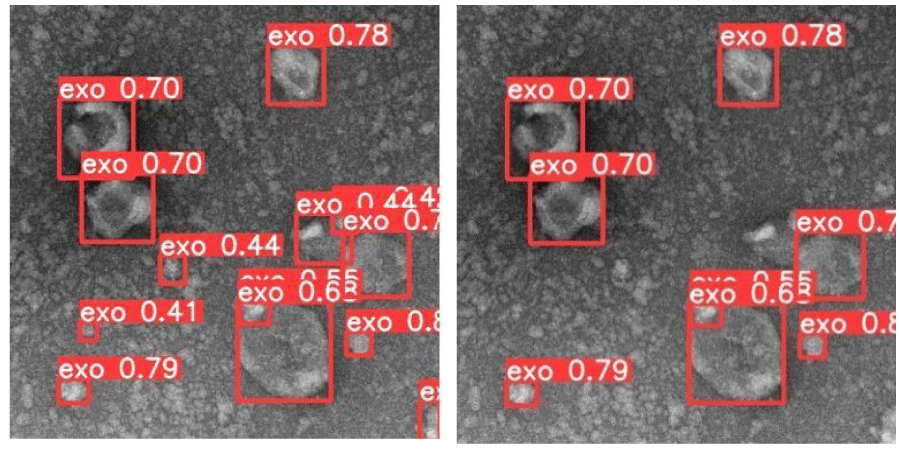
A score reflecting the confidence assigned to the detection of each object can be used as a parameter in the detector to limit the selection to high confidence object detections if relevant, like demonstrated in Figure 3.
The algorithm can now be fine-tuned through the parameters available for inference and tested in the remaining images to which we have ground truth data. This 40 images dataset has the corresponding 40 annotated images.
Conclusion
The possibilities of deep learning algorithms such as YOLO v5 seem to cover the void caused by procedures outputting high volumes of data at the end of the processing chain when huge volumes of data need to be processed and human labor availability is limited.
A fine tunned algorithm can take days to train and test, but the time of inference will be very short compared to the need of a human operator, and the results are consistent and robust if the criteria for the train were studied for that case and correctly applied. As the training focus on the attributes of the new objects of interest, transfer learning can be achieved with CNNs that had been trained with large data sets, because the basic attributes like edges and contrast differences are common to all objects and are at the base of the first layers, the details concerning our objects will be added to an already robust decision procedure to refine it. This kind of systematic approach can be achieved with open-source software and a current laptop. In future work, we’ll implement the algorithm with TEM datasets of other biological particles of interest, such as polyomavirus and adenovirus.
We want to thank Karel Stepka from the Centre for Biomedical Image Analysis, Masaryk University, Czech Republic who made this data set available in 2016.
- Dilsiz N (2020) Role of exosomes and exosomal microRNAs in cancer. Future Sci OA 6: FSO465. Link: https://bit.ly/3stFBYZ
- Xitong D, Xiaorong Z (2016) Targeted therapeutic delivery using engineered exosomes and its applications in cardiovascular diseases. Gene 575: 377-384. Link: https://bit.ly/33SBQlL
- Chen H, Wang L, Zeng X, Schwarz H, Nanda HS, et al. (2021) Exosomes, a New Star for Targeted Delivery. Front Cell Dev Biol 9: 751079. Link: https://bit.ly/3qCWmyD
- Théry C (2011) Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 3: 15. Link: https://bit.ly/3FrgslA
- Tamkovich SN, Tutanov OS, Laktionov PP (2016) Exosomes: generation, structure, transport, biological activity and diagnostic application. Membrane Cell Biol 10: 163-173. Link: https://bit.ly/3JgFDJF
- Zabeo D, Cvjetkovic A, Lässer C, Schorb M, Lötvall J, et al. (2017) Exosomes purified from a single cell type have diverse morphology and composition, J Extracell Vesicles 20: 1329476. Link: https://bit.ly/3FomBii
- Gutha S (2021) Object Detection Algorithm — YOLO v5 Architecture. Analytics Vidhya (medium.com). Link: https://bit.ly/3FzHLKc
- Jocher G (2021) GitHub. Link: https://bit.ly/30Rt862
- Make Sense. Link: https://bit.ly/3JnWZVg
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
